As an ophthalmologist, you’re often on the front line when it comes to diagnosing a patient with uveal melanoma. When this diagnosis is made, it’s often accompanied by a flood of questions from the patient, such as: What’s the prognosis? Did it spread anywhere else? For years, ophthalmologists have relied on UM classification systems to help answer these questions. Recently, a new classification system, The Cancer Genome Atlas, was developed, and bases its classifications on a tumor’s genetic profile. Here, we present an overview of the genetic aberrations that define this classification system, as well as a review of the literature for outcomes in patients with uveal melanoma stratified by this system.
Classifying Uveal Melanoma
Uveal melanoma, the most common primary intraocular malignancy in adults, is a malignant neoplasm affecting the iris, ciliary body and choroid.1 While these tumors can be visually threatening, they also carry a significant risk for metastatic disease with the most common sites being the liver, lung, bone and skin, respectively.2 As mentioned earlier, ophthalmologists are usually the first to diagnose UM and refer a patient to an ocular oncologist. Making an accurate prognosis and determining the risks for systemic metastasis depends in large part on tumor size, location, genetics and classification.3
There are a few classification systems for UM that aid in the prediction of prognosis. These systems have taken into consideration the anatomic features of the tumor and/or genetic aberrations characterizing the tumor.3-9 The American Joint Committee on Cancer Classification 8th edition criteria separated UM into categories and stages based on the tumor anatomy, with a focus on involvement of the choroid and ciliary body, basal dimension and thickness of the tumor, distance to the foveola, any documentation of tumor growth and evidence for extraocular extension.3
The Cancer Genome Atlas
The Cancer Genome Atlas (TCGA) classification is the result of an international collaboration led by the National Cancer Institute Center for Cancer Genomics and the National Human Genome Research Institute in 2005. TCGA project’s mission was to characterize the molecular changes that occur in cancer cells by analyzing data collected from thousands of human samples. All told, TCGA collected and analyzed over 20,000 tissue samples from 33 cancer types (including those specific to uveal melanoma) to elucidate not only the genes that play a role in each type of cancer, but also identify protein-complexes and pathways that contribute to these malignancies.10-12 This analysis led to refined systems of grouping for various cancers, allowing treatment plans to be modified to address the clinical, histopathologic, and now genetic, alterations in cancer types. More than 2.5 petabytes (2.5 million gigabytes) of data relating to the genetic and proteomic characteristics of these cancers have been generated by this analysis.13
 |
|
Abbreviations: EIF1AX, Eukaryotic Translation Initiation Factor 1A X-Linked; SF3B1, Splicing Factor 3b Subunit 1; BAP1, BRCA1 Associated Protein 1 Adapted from: Robertson AG, Shih J, Yau C, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:2:204-220; Jager MJ, Brouwer NJ, Esmaeli B. The Cancer Genome Atlas Project: An integrated molecular view of uveal melanoma. Ophthalmology 2018;125:8:1139e1142. |
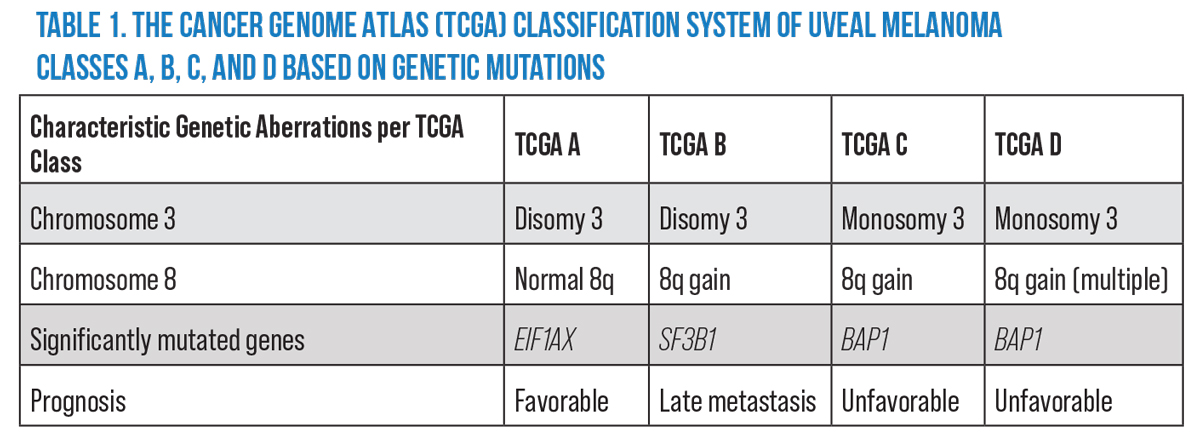
The TCGA researchers identified a multitude of genetic, histologic, immune and proteomic influences in uveal melanoma which contribute to the disease’s prognosis.11 Based on their findings, the researchers11,12 synthesized a classification system that stratifies uveal melanoma into four groups: TCGA Group A (Figure 1A); TCGA Group B (Figure 1B); TCGA Group C (Figure 1C); and TCGA Group D (Figure 1D). Each TCGA group reliably characterizes uveal melanomas by severity of disease and risk for metastasis (Table 1).12
TCGA Findings in Uveal Melanoma
In 2017, A. Gordon Robertson, PhD, and co-workers at Vancouver’s Michael Smith Genome Sciences Centre first published TCGA data in the Rare Tumor Project collected from 80 human eyes with UM and extrapolated on the relevant genetic pathways involved. The researchers found four molecular and clinical distinct subsets of UM.11 A year later, The Netherlands’ Martine J. Jager, MD, and colleagues simplified the findings and proposed the four-category classification system of UM that’s been implemented clinically to predict prognostic characteristics of the disease, such as rate of metastasis and death.12 The four categories each differ genetically based on chromosome 3 and 8 findings.11,12 This classification system has since been documented to be more accurate than the AJCC 8th edition regarding its use in prediction of five-year rate of metastasis.3
TCGA explored numerous alterations in DNA, RNA expression, protein translation, methylation status and immunologic factors, and ultimately identified four clinically relevant subtypes of UM, each with distinct genetic aberrations and prognostic characteristics. These genetic mutations not only correlated with clinical outcomes of UM, but they were mutations specifically not found in cutaneous melanoma. The primary factor in deciding prognostic outcomes is the status of chromosome 3: Disomy 3 (D3) showed favorable prognosis while monosomy 3 (M3) showed unfavorable prognosis. Within each grouping of D3 and M3 were additional stratifications decided by absence or presence of functional gains in 8q.11
Among the D3 tumors, Dr. Robertson identified two genetic aberrations—nearly mutually exclusive—that contributed to tumor pathogenesis. Mutations in EIF1AX were identified in uveal melanomas that were characterized by chromosome 3 disomy and no gains in 8q (D3, no 8q gain).11 Mutations in SF3B1 were also identified in disomy 3 tumors, but only if they had partial functional gains in 8q (D3, partial 8q gain). These two D3 tumor types had distinct somatic copy number alterations from one another, leading to D3 tumors being subclassified into tumors with D3 and no 8q gain and tumors with D3 and 8q gain (Table 1).11
The UMs which showed M3 demonstrated loss of function of tumor suppression gene BAP-1 (located on 3p21). Tumors with this genetic aberration were followed by a global DNA methylation state and were highly correlated with UM metastasis. Even though the global methylation state was shown to be shared among all M3 tumors, the authors identified differences in cell signaling pathways and protein expression as well as differences in clinical outcomes between M3 tumors without multiple 8q gains (M3, 8q gain) and the M3 tumors with multiple 8q gains (M3, multiple 8q gains). Tumors with multiple 8q gains showed evidence of presence of 8q isochromosomes (chromosome 8 with 2 q arms) (Table 1).11
Based on the findings of Dr. Robertson’s group, Dr. Jager and her colleagues proposed TCGA Group A (D3, no 8q gain), TCGA Group B (D3, partial 8q gain) TCGA Group C (M3, 8q gain), and TCGA Group D (M3, multiple 8q gains).11,12 This classification system has been widely used clinically, and has been the basis for important validation studies and studies focused on the multitude of clinical correlations and implications of this classification system.1,14,15 Moreover, TCGA’s classification system has been shown to improve the level of predictive precision compared to the previously dominant American Joint Committee on Cancer classification system when both TCGA and AJCC criteria were taken into consideration.16
 |
|
Adapted from: Shields CL, Mayro EL, Bas Z, et al. Ten‑year outcomes of uveal melanoma based on The Cancer Genome Atlas (TCGA) classification in 1001 cases. Indian J Ophthalmol 2021;69:1839-45. *Metastasis elsewhere includes metastasis to bone, brain, breast, intestine, distant lymph nodes, mesentery, muscle and skin. |
The Current Literature on TCGA and UM
Since the publication of the TCGA’s classification system, efforts to apply the system clinically have revealed a great deal about patient prognosis in patients in increasing TCGA categories (A vs. B vs. C vs. D). In 2019, a group of researchers was the first to apply the TCGA classification clinically to a cohort of 658 patients with UM, primarily focusing on the difference in rates of metastasis and death at five years.1 In this cohort, the researchers found that not only did the rate of metastasis increase with increasing TCGA group (3 vs. 10 vs. 25 vs. 41 percent, respectively; p<0.001), but the mean time to metastasis decreased with increasing TCGA group (42.1 vs. 41 vs. 30.8 vs. 21.1 months, respectively; p<0.001).
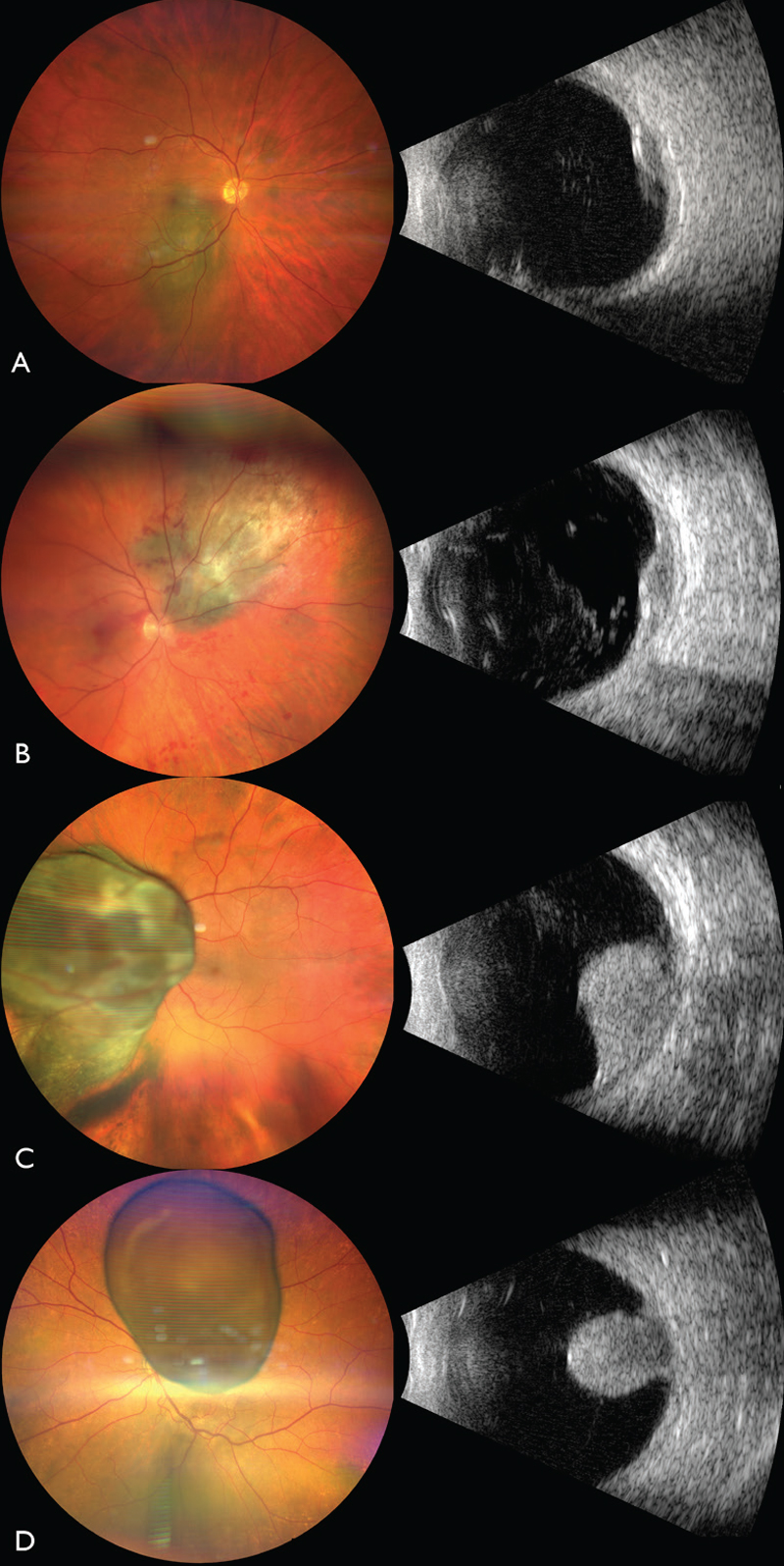 |
| Figure 1. The Cancer Genome Atlas classification of uveal melanoma using fine needle aspiration biopsy. (A) TCGA Group A UM, measuring 11 mm x 7 mm in base with shallow subretinal fluid and orange pigment (left panel) and ultrasound showing an acoustically-hollow tumor with thickness of 3.1 mm (right panel). (B) TCGA Group B UM, measuring 12 mm x 11 mm in base with shallow subretinal fluid (left panel) and ultrasound showing an acoustically-hollow tumor with a thickness of 3.9 mm (right panel). (C) TCGA Group C UM overhanging the optic disc, measuring 18 mm x 17 mm in base with shallow subretinal fluid (left panel) and ultrasound showing a mushroom-shaped tumor with thickness of 10.7 mm (right panel). (D) TCGA Group D UM overhanging the optic disc, measuring 14 mm x 10 mm in base with shallow subretinal fluid and overlying retinal invasion (left panel) and ultrasound showing a mushroom-shaped tumor with thickness of 10.3 mm (right panel). |
When types of metastasis were analyzed individually, the rates of metastasis similarly increased with increasing TCGA grouping with statistical significance. This was shown for liver metastasis (2 vs. 10 vs. 24 vs. 40 percent, respectively; p<0.001), lung metastasis (<1 vs. 1 vs. 3 vs. 7 percent, respectively; p<0.001), as well as a grouping of other types of metastasis, including bone, brain, breast, intestine, distant lymph nodes, mesentery, muscle and skin (1 vs. 3 vs. 3 vs. 9 percent, respectively; p=0.001). Other statistically significant associations with increasing TCGA grouping were also described, including lower visual acuity, more anterior tumor location, lower frequency of tumor epicenter in the choroid, greater distance from the optic nerve and foveola, greater basal diameter and greater tumor thickness as TCGA group increased.1
One of this article’s authors, Carol Shields, MD, and her group then performed a follow-up study to further validate these findings on a cohort of 1,001 eyes with a follow-up period of 10 years.14 Unsurprisingly, this study confirmed an increasing rate of metastasis with increasing TCGA classification (3 vs. 9 vs. 20, vs. 46 percent, respectively; p<0.001) as well as decreased time interval to metastasis (37.4 vs. 38.7 vs. 27.7 vs. 21.5 months, respectively; p=0.009).
The rates of individual types of metastasis similarly increased with increased TCGA grouping. This was shown in liver metastasis (2 vs. 9 vs. 20 vs. 46 percent, respectively; p<0.001), lung metastasis (<1 vs. 1 vs. 4 vs. 10 percent, respectively; p<0.001), and other metastasis (1 vs. 4 vs. 5 vs. 14 percent, respectively; p<0.001).
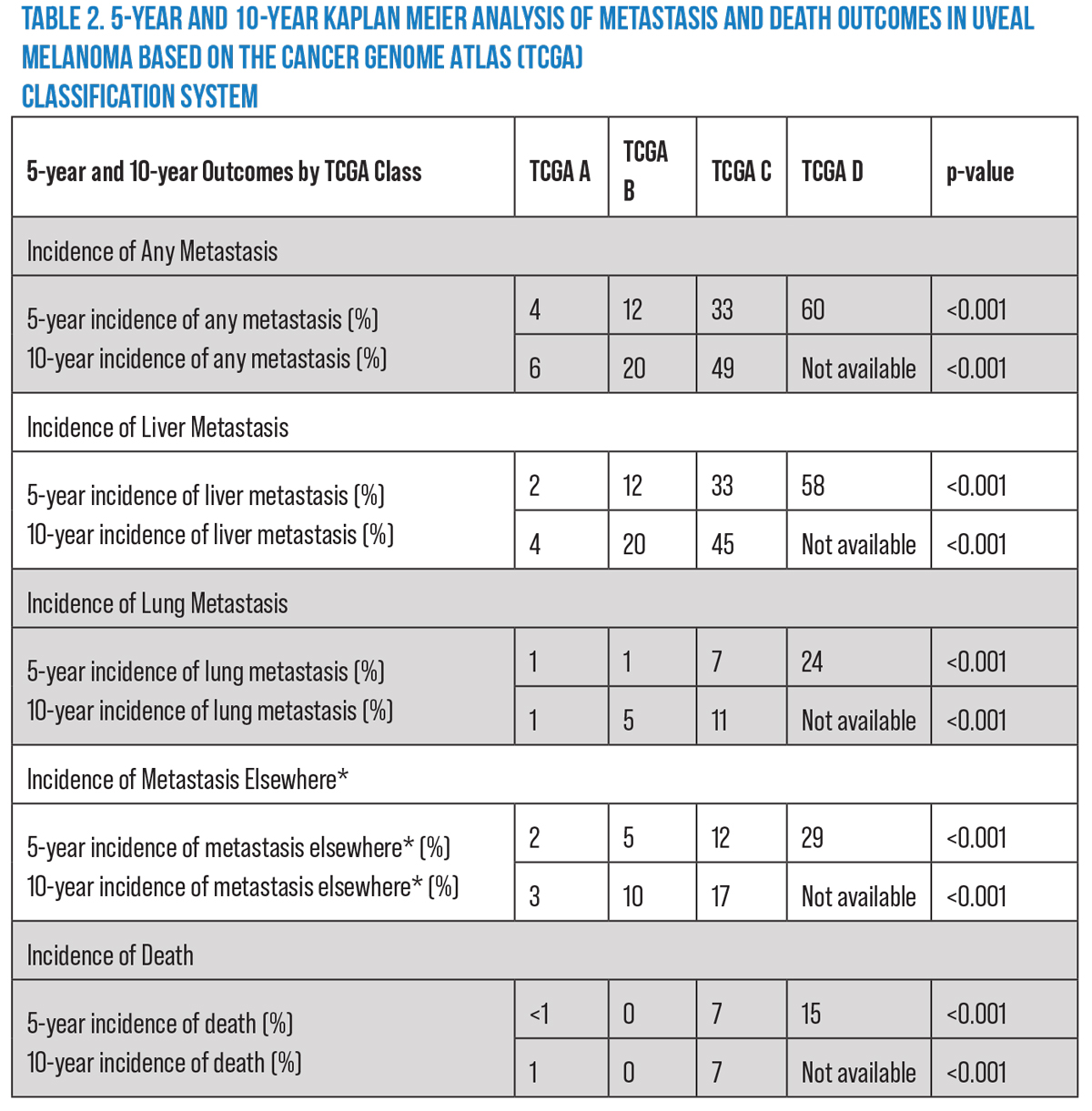
Dr. Shields’ group also found that the rates of melanoma-related death increased with increasing TCGA category (<1 vs. 0 vs. 2 vs. 7 percent, respectively; p=0.003). Kaplan Meier analysis of this cohort demonstrated increasing metastasis rates at the five-year mark (4, 12, 33, and 60 percent, respectively; p<0.001) as well as the 10-year mark (6, 20 and 49 [data on the last category isn’t available], respectively; p<0.001).14 This data has been very helpful for counseling patients, managing their expectations and coordinating their treatment (Table 2).
A collaboration between Wills Eye Hospital and Leiden University Medical Center in the Netherlands then explored the impact of TCGA grouping on patient outcomes based on iris color; lighter iris color (blue, gray, and green irides) has been shown to be an independent factor increasing the risk for development of UM compared to darker irises.15 They found that although there was no difference in mortality when stratified by iris color (p=0.28), the chromosome 3 and 8q copy numbers made a greater impact on survival in patients with lighter irides (gray, blue, and green) as well as in patients with lightly pigmented tumors.
Chromosome 3 status impacted patient survival in blue-iris (p=0.001) and green-iris patients (p<0.0001), but not in brown-iris patients (p=0.43). The same was found to be true for the impact of 8q status in blue-iris (p=0.001) and green-iris patients (p<0.001) when compared to its effect on survival in brown-iris patients (p=0.28). They concluded that iris pigmentation likely plays a role in the oncogenic behavior of UM tumors and that iris color shouldn’t be overlooked when assessing patients with UM.15
Efforts to further refine this system of UM classification have taken into account the system that TCGA seemed to be replacing, the AJCC. Unlike the TCGA criteria, AJCC criteria include clinical factors of the tumor, such as tumor size, ciliary body involvement and extrascleral extension.3 A group at Leiden University Medical Center led by Maria Chiara Gelmi, MD, found that combining the AJCC with the TCGA criteria provided a refined system with increased prognostic accuracy compared to either system alone. They essentially found that chromosomal aberrations had a greater impact on AJCC stage II and III tumors compared to AJCC stage I tumors. They also found mutually exclusive prognostic differences in TCGA groups C and D that were further subclassified into AJCC stage II and stage III. Thus, they concluded that TCGA C patients should also have clinical factors such as tumor size, involvement of the ciliary body and extrascleral extension (all predictive of a poorer prognosis) taken into consideration. They also found that chromosome status should be taken into account when estimating a prognosis with tumors staged as AJCC II or III.16
Subsequently, Viktor Gill, MD, PhD, and colleagues at the Department of Pathology, Västmanland Hospital in Västerås, Sweden, expanded on the idea that genetic and clinical factors that hold predictive value independent of the genetic status should be combined when categorizing patients with UM.17 They explored the combination of clinical and genetic prognostic factors in 1,796 patients with UM from Wills Eye Hospital and St. Erik Eye Hospital in Stockholm.17 A multivariate Cox regression model determined that male sex, patient age at diagnosis, AJCC T category, monosomy 3 and tumor involvement in the ciliary body were all independent predictors of metastasis. Using this data, they constructed and validated a point system (0-12.5) within their patient cohort. Points were assigned as follows when 8q data was available:
• 0.5 points for male sex;
• 0.5 points for age over 70 years;
• 0.5 points for ciliary body involvement;
• 1.5 points for extrascleral extension of ≤5mm;
• 3 points for extrascleral extension >5mm;
• 2 points for monosomy 3;
• 1 point for 3 copies of 8q;
• 2 points for >3 copies of 8q;
• 1 point for AJCC-T category 1;
• 2 points for AJCC-T category 2;
• 3 points for AJCC-T category 3; and
• 4 points for AJCC-T category 4.
The resulting total determines whether the patient belongs to prognostic group 1 (2 points or fewer), prognostic group 2 (2.5 to 4.5 points), prognostic group 3 (5 to 7 points), or prognostic group 4 (7.5 points and up). Their univariate and multivariate regression models suggested a more accurate predictability of mortality related to UM than AJCC or TCGA individually.17
In summary, TCGA’s classification system has provided a highly reliable and applicable prognostication system for patients with UM, guiding expectations for morbidity and mortality as well as approaches to treatment and coordinated systemic monitoring. The system has been widely used in ocular oncology and serves as a reliable point of reference for projects aiming at improving how UM is categorized. Overall, there’s definite value in taking patient genetics and clinical data into account when caring for UM patients. Further research on larger patient cohorts may be done to determine more exactly the effect each individual factor has on a patient’s prognosis.
Support provided in part by the Eye Tumor Research Foundation, Philadelphia (CLS). The funders had no role in the design or conduct of the study; the collection, analysis and interpretation of the data; or the preparation, review or approval of the manuscript. Dr. Shields has had full access to all the data in the study and takes responsibility for its integrity.
Inquiries: Carol L. Shields, MD, Ocular Oncology Service, 840 Walnut Street, Suite 1440, Philadelphia, PA 19107. Tel: (215) 928-3105, Fax: (215) 928-1140, Email: carolshields@gmail.com
Dr. Regillo is the director of the Retina Service of Wills Eye Hospital, a professor of ophthalmology at Thomas Jefferson University School of Medicine and the principle investigator for numerous major international clinical trials.
Dr. Yonekawa is an assistant professor of ophthalmology at Sidney Kimmel Medical College at Thomas Jefferson University. He serves on the Education Committee of the American Society of Retina Specialists and on the Executive Committee for the Vit Buckle Society, where he is also the vice president for academic programming.
Mr. Kevin Card is a medical student at the University of Hawai’i John A. Burns School of Medicine currently doing a one-year research fellowship at the Ocular Oncology Service at Wills Eye Hospital.
Dr. Konstantinou is a clinical ocular oncology fellow at Wills Eye Hospital.
Dr. Shields is chief of the Ocular Oncology Service at Wills Eye Hospital and professor of ophthalmology at Thomas Jefferson University in Philadelphia. None of the authors has any conflicts of interest to report.
1. Vichitvejpaisal P, Dalvin LA, Mazloumi M, Ewens KG, Ganguly A, Shields CL. Genetic analysis of uveal melanoma in 658 patients using the cancer genome atlas classification of uveal melanoma as A, B, C, and D. Ophthalmology 2019;126:10:1445-1453.
2. Diener-West M, Reynolds SM, Agugliaro DJ, et al. Development of metastatic disease after enrollment in the COMS trials for treatment of choroidal melanoma: Collaborative Ocular Melanoma Study Group Report No. 26. Arch Ophthalmol 2005;123:12:1639-1643.
3. Mazloumi M, Vichitvejpaisal P, Dalvin LA, Yaghy A, Ewens KG, Ganguly A, et al. Accuracy of the cancer genome atlas (TCGA) classification versus American joint committee on cancer (AJCC) classification for prediction of metastasis in patients with uveal melanoma. JAMA Ophthalmol 2020;138;260‑7.
4. Dogrusoz M, Jager MJ. Genetic prognostication in uveal melanoma. Acta Ophthalmol 2018;96:331‑47.
5. Vaquero‑Garcia J, Lalonde E, Ewens KG, et al. PRiMeUM: A model for predicting risk of metastasis in uveal melanoma. Invest Ophthalmol Vis Sci 2017;58:4096‑105.
6. Dogrusoz M, Jager MJ, Damato B. Uveal melanoma treatment and prognostication. Asia Pac J Ophthalmol (Phila) 2017;6:186‑96.
7. Ewens KG, Kanetsky PA, Richards‑Yutz J, et al. Genomic profile of 320 uveal melanoma cases: chromosome 8p‑loss and metastatic outcome. Invest Ophthalmol Vis Sci 2013;54:5721‑9.
8. Damato B, Dopierala JA, Coupland SE. Genotypic profiling of 452 choroidal melanomas with multiplex ligation‑dependent probe amplification. Clin Cancer Res 2010;16:6083‑92.
9. Damato B, Coupland SE. Translating uveal melanoma cytogenetics into clinical care. Arch Ophthalmol 2009;127:423‑9.
10. Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, et al. The cancer genome atlas pan‑cancer analysis project. Nat Genet 2013;45:1113‑20.
11. Robertson AG, Shih J, Yau C, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 2017;32:204‑20.e15.
12. Jager MJ, Brouwer NJ, Esmaeli B. The cancer genome atlas project: An integrated molecular view of uveal melanoma. Ophthalmology 2018;125:1139‑42.
13. National Institute of Health, National Cancer Institute. The cancer genome atlas project. https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga. Accessed August 1, 2022.
14. Shields CL, Mayro EL, Dockery PW, et al. Ten-year outcomes of uveal melanoma based on the cancer genome atlas (TCGA) classification in 1001 Cases. Indian J Ophthalmol 2021;69:7:1839-1845.
15. Wierenga APA, Brouwer NJ, Gelmi MC, et al. Chromosome 3 and 8q aberrations in uveal melanoma show greater impact on survival in patients with light iris versus dark iris color. Ophthalmology 2022;129:4:421-430.
16. Gelmi MC, Bas Z, Malkani K, Ganguly A, Shields CL, Jager MJ. Adding the cancer genome atlas chromosome classes to American joint committee on cancer system offers more precise prognostication in uveal melanoma. Ophthalmology 2022;129:4:431-437.
17. Gill VT, Sabazade S, Herrspiegel C, et al. A prognostic classification system for uveal melanoma based on a combination of patient age and sex, the American joint committee on cancer and the cancer genome atlas models. Acta Ophthalmol 2022 Jul 8. doi: 10.1111/aos.15210. Online ahead of print.



